Are those genes in my Tetris?
Scientists find new digital ways to decipher genomic data
Amy Nordrum • November 13, 2013
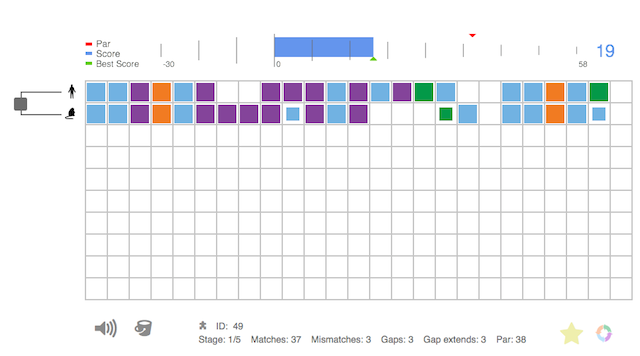
A screenshot of Phylo, a computer game. Stacking the colored boxes in matching columns helps researchers sort genetic data. [Image Credit: http://phylo.cs.mcgill.ca]
So much data, so little time. That’s the problem confronting geneticists studying the bewilderingly complex human genome, in which no fewer than three billion – yes, with a ‘b’ – possible combinations of base pairs (think A,T,C,G) comprise about 20,500 genes.
That deluge of data creates a new challenge — how to analyze all those genes in the search to identify disease or traits? Scientists are finding some clever digital solutions. A group of 300 researchers is storing and sharing their genomic data in the cloud, while another geneticist has debuted the open-source version of a computer game called Phylo (inspired by Tetris), where users playing with patterns inadvertently clean up messy data for researchers.
The cloud technology that many people use to upload and edit in Google drive will now enable genome scientists to share their data across servers, too. Jeffrey Reid, a geneticist from the Baylor College of Medicine’s Human Genome Sequencing Center has teamed up with Amazon Web Services and DNAnexus, a company that builds digital storage for researchers, to develop an online database and integrated user network to share data of 14,000 individuals mapped by his colleagues—a set large enough that researchers with limited data storage and network speed were having trouble accessing it remotely.
“Your instinct as a scientist is to want to physically have your data but at some point when the data set becomes so big and the analysis so unwieldy, it’s just not practical,” Reid said.
The 300 scientists with access to the cloud will use it to run analyses for the CHARGE project, a research group primarily focused on finding mutations that cause heart disease and aging.
“We want to understand how the genome affects everything in health,” Reid said. “To do that, we have to create a large sample set and ask multiple questions at the same time.”
The developers announced the new system Oct. 25 at the annual meeting of the American Society of Human Genetics in Boston. Reid says it is a sign of the times, when data crunching technology is rapidly evolving.
“Seven years ago, it was very difficult to sequence a genome and now you can do it within days,” Reid said. “Bringing that together with this revolution in computing in hardware and software is going to change medicine.”
Amateur scientists who don’t have access to a cloud database but still want to play with genes can get their fix with Phylo, an online computer game. In Phylo, players arrange colored boxes into patterns of rows and columns, much like Tetris. But in this version, players are technically sorting by multiple sequence alignment—the term for the way similar sections on different genes typically line up.
“I was trying to think, ‘What was the most useful problem you could solve to advance genetic research?’” said Jérôme Waldispühl, the developer and a computer scientist at McGill University. “The first thing everybody is doing is applying multiple sequence alignment.”
Players are actually fixing errors made by a computer program in its first run through the data because humans are better than robots at recognizing certain types of patterns. Correcting the alignment helps researchers compare and contrast the genes, and pick out mutations.
In three years, 300,000 users have played the game. Waldispühl created Phylo to identify genes associated with common diseases. At just over a year after the launch, players had proposed 350,000 corrections to researchers studying diabetes, cancer and tumors. These adjustments helped improve 521 gene segments in that time.
“So far, it works. Because what you’re doing with Phylo is not the research from A-Z where you’re trying to find a whole scientific discovery,” Waldispühl said. “You’re providing the data for genetic researchers to make advances.”
Waldispühl is now turning the game’s users loose on genetic data worldwide. He just announced that he will offer up Phylo to other researchers interested in using it for a project. He will link the game’s players to new data sets and put players to work on sorting genes specific to any study. This version is called Open-Phylo. Waldispühl tested the program by asking players to work with three cancer genes and found that they “consistently improve the alignments,” as stated in a research paper in Genome Biology.
The CHARGE project and Open Phylo will make it easier and more efficient to sort data, but there is still plenty of room for more projects to make a digital dent in genetic analysis. The greatest potential for breakthroughs in the field may now rest in finding the best ways to make sense of our data-rich genome.
Reid, at least, is enjoying the limitless potential.
“I don’t think people realize how unique a moment this is in the science of genomics,” he said. “This is the coolest thing I could possibly be doing right now, so I’m really happy.”
*Correction, November 14, 2013:
Originally, this article read that “players had fixed 350,000 misalignments for researchers” in its first year. This statement is misleading. Rather, players had proposed 350,000 solutions by this time.