Viral genome built in a test tube
A new method to stitch together DNA can assemble a virus genome in one day
Niko McCarty • November 17, 2020
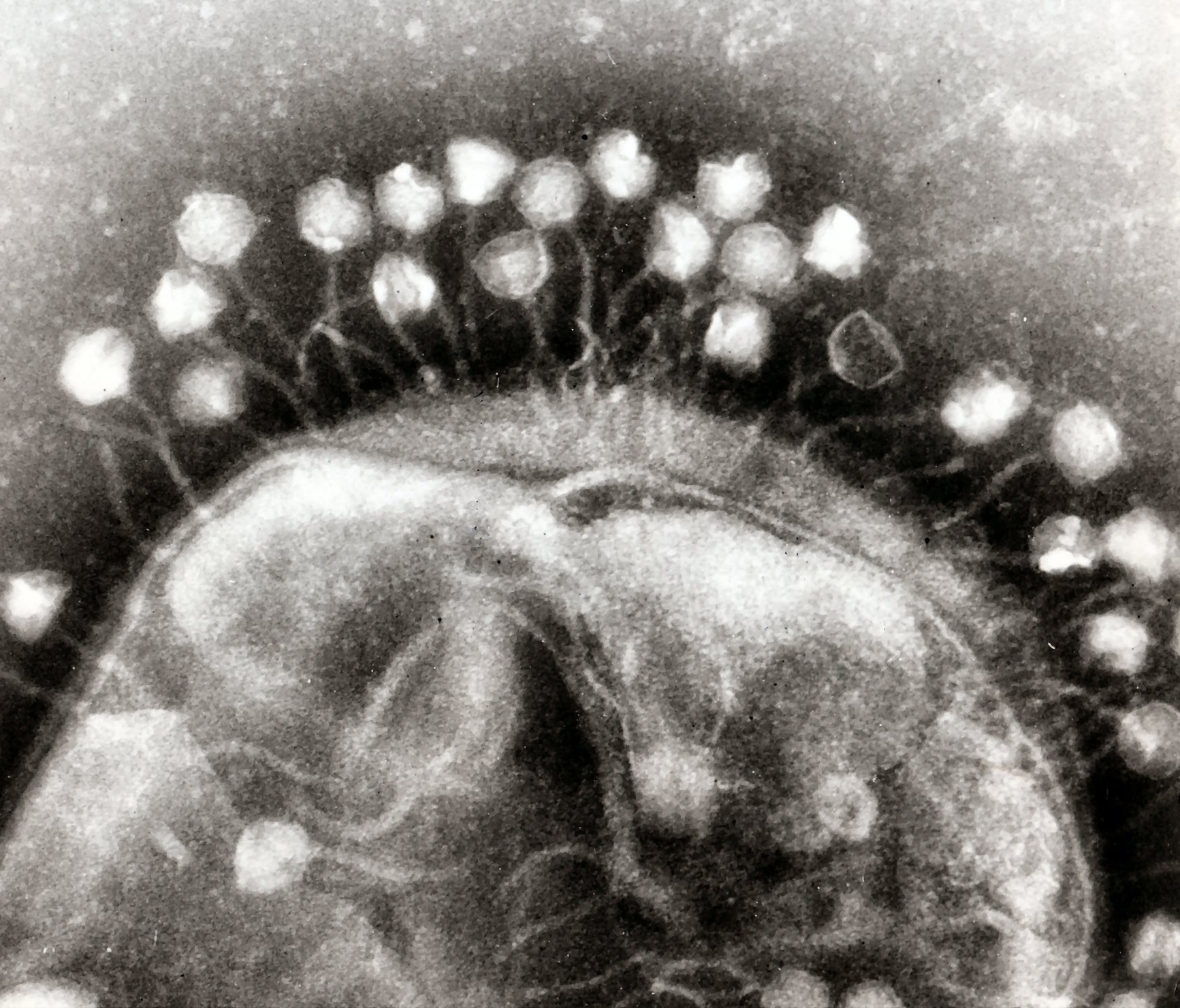
Bacteriophages cling to the outer membrane of a bacterial cell. Image taken with a transmission electron microscope. [Credit: Dr. Graham Beards at Wikimedia | CC BY-SA 3.0]
To understand how genomes work, some scientists design and build new ones — a process that can take months or years. Now, an improved method has drastically increased the number of DNA pieces that can be stitched together, opening the door for small, designer genomes built in a single day.
In 2010, the J. Craig Venter Institute reported the first self-replicating, synthetic bacterial cell. In that case, the organism’s genome was created with chemistry; its DNA was joined together slowly, piece by piece. That project cost millions of dollars and took several years to complete, but it helped researchers better understand how DNA sequences, and the order of genes, affect the way an organism looks or behaves.
Ten years on, it’s easier than ever to build synthetic genomes. In a new study, researchers at New England Biolabs, a private company in Ipswich, Massachusetts, assembled 35 distinct fragments of DNA in one test tube. That’s a big improvement over existing methods for joining together DNA, which were typically limited to a handful of genetic fragments. The results were published in September in the journal PLOS ONE. But the researchers behind this work tell me that their method is already outdated.
The same team has already assembled an entire bacteriophage genome in a test tube from 50 distinct pieces of DNA. That’s according to Greg Lohman, a senior scientist at New England Biolabs and lead author of the initial study in PLOS ONE. These new results were revealed Sept. 30 at the virtual SynBioBeta conference.
A bacteriophage is a type of virus that infects bacteria, and the genome assembled by Lohman’s team contains 40,000 nucleotides, or letters; by comparison, the human genome has over 3 billion letters. Still, the small genome is an impressive start.
The method used by Lohman’s team to build the bacteriophage genome is called Golden Gate Assembly. It works like this: Molecular machines, called proteins, chew up the ends of each DNA piece. The proteins leave behind “sticky” ends, which can then be joined to other pieces with complementary ends. To join 50 pieces together, the researchers meticulously tested, tweaked, and tuned those sticky ends until they identified an optimal combination.
Building a synthetic bacteriophage genome is just a proof of concept for now, but the underlying method that made this achievement possible could be used to create any number of small, designer genomes. Several synthetic biologists agree that Lohman’s research is significant. Just a few years ago, assembling DNA fragments was complicated, even for experts.
“When I was a first year PhD student, one of my rotation projects was to make a construct with three [DNA fragments],” said Naomi Nakayama, a researcher at Imperial College London who was not involved in the study. “I worked on it for three months. I could only put in two [pieces] in the end.”